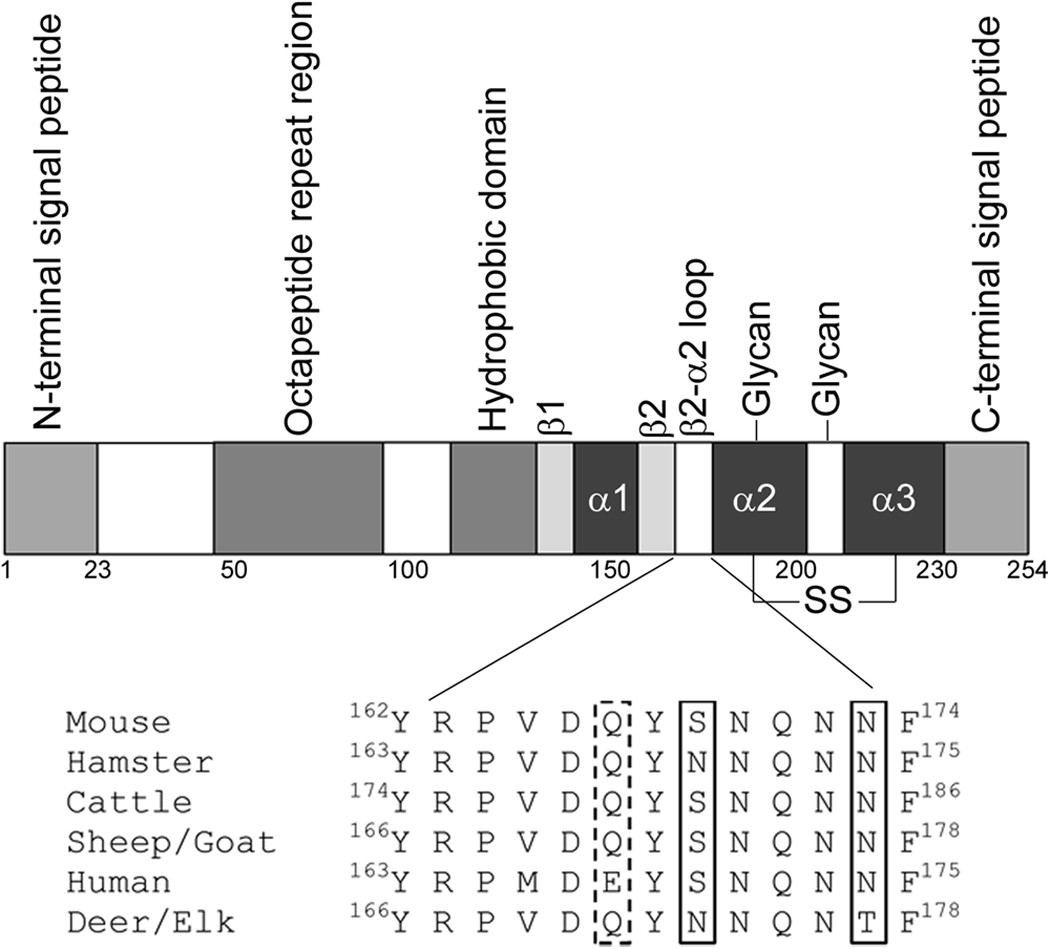
Complex folding and misfolding effects of deer-specific amino acid substitutions in the β2-α2 loop of murine prion protein | Scientific Reports
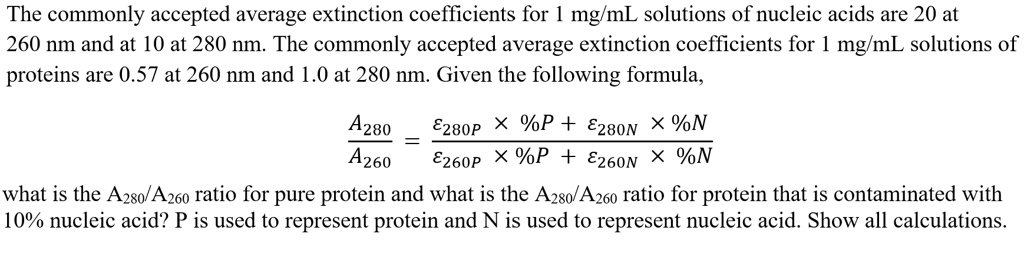
SOLVED: The commonly accepted average extinction coefficients for mg/mL solutions of nucleic acids are 20 at 260 nm and at 10 at 280 nm The commonly accepted average extinction coefficients for mglmL

Molar absorption coefficients for Trp, Tyr, and cystine based on an... | Download Scientific Diagram
Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. - Abstract - Europe PMC

Observed and predicted molar absorption coefficients at 280 nm for 80... | Download Scientific Diagram

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal

Molecules | Free Full-Text | Evaluation of Peptide/Protein Self-Assembly and Aggregation by Spectroscopic Methods