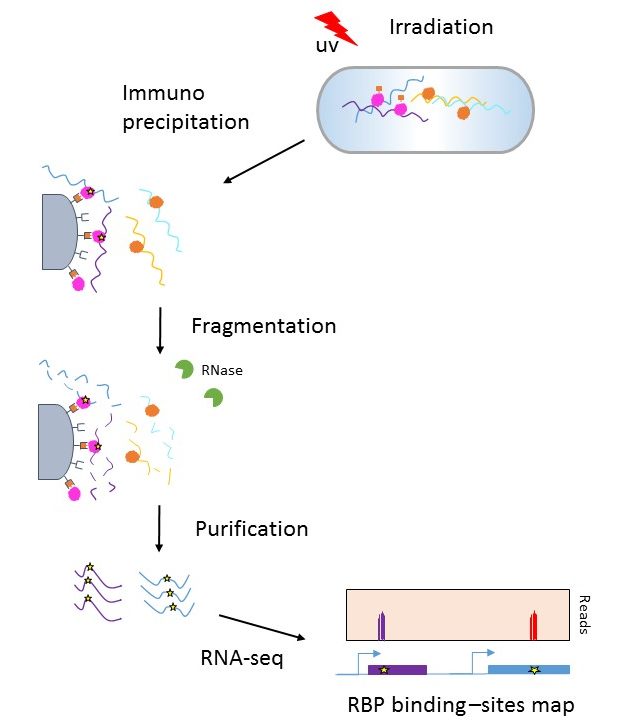
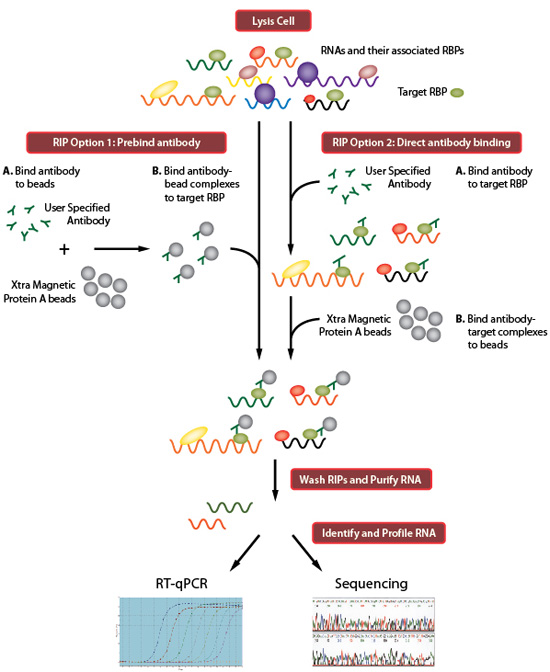
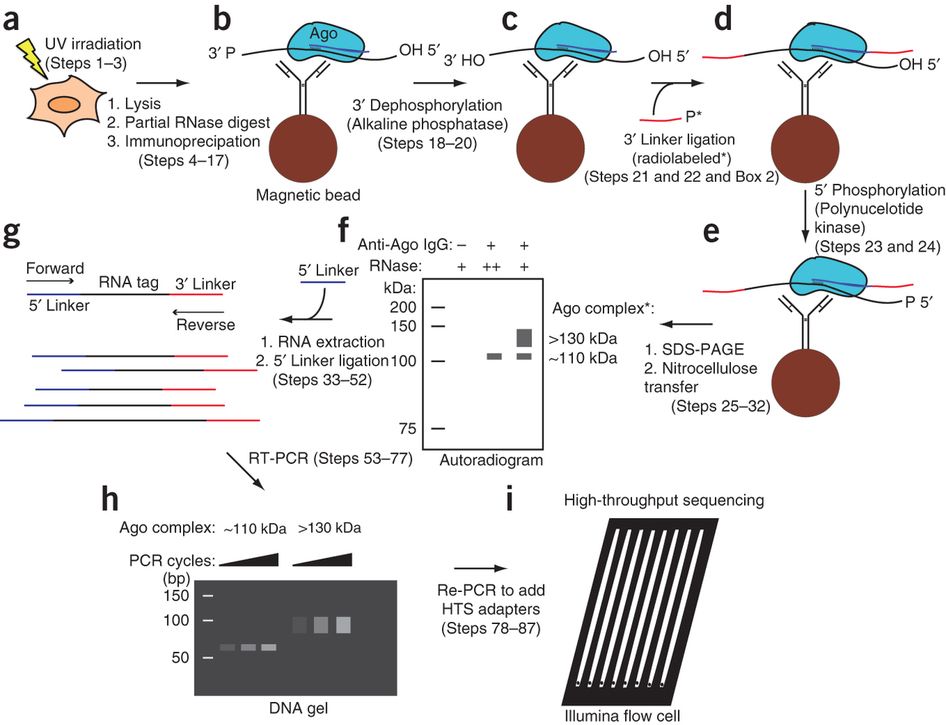
Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP: Cell

RNA structure, binding, and coordination in Arabidopsis - Foley - 2017 - WIREs RNA - Wiley Online Library
The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid | PLOS Pathogens
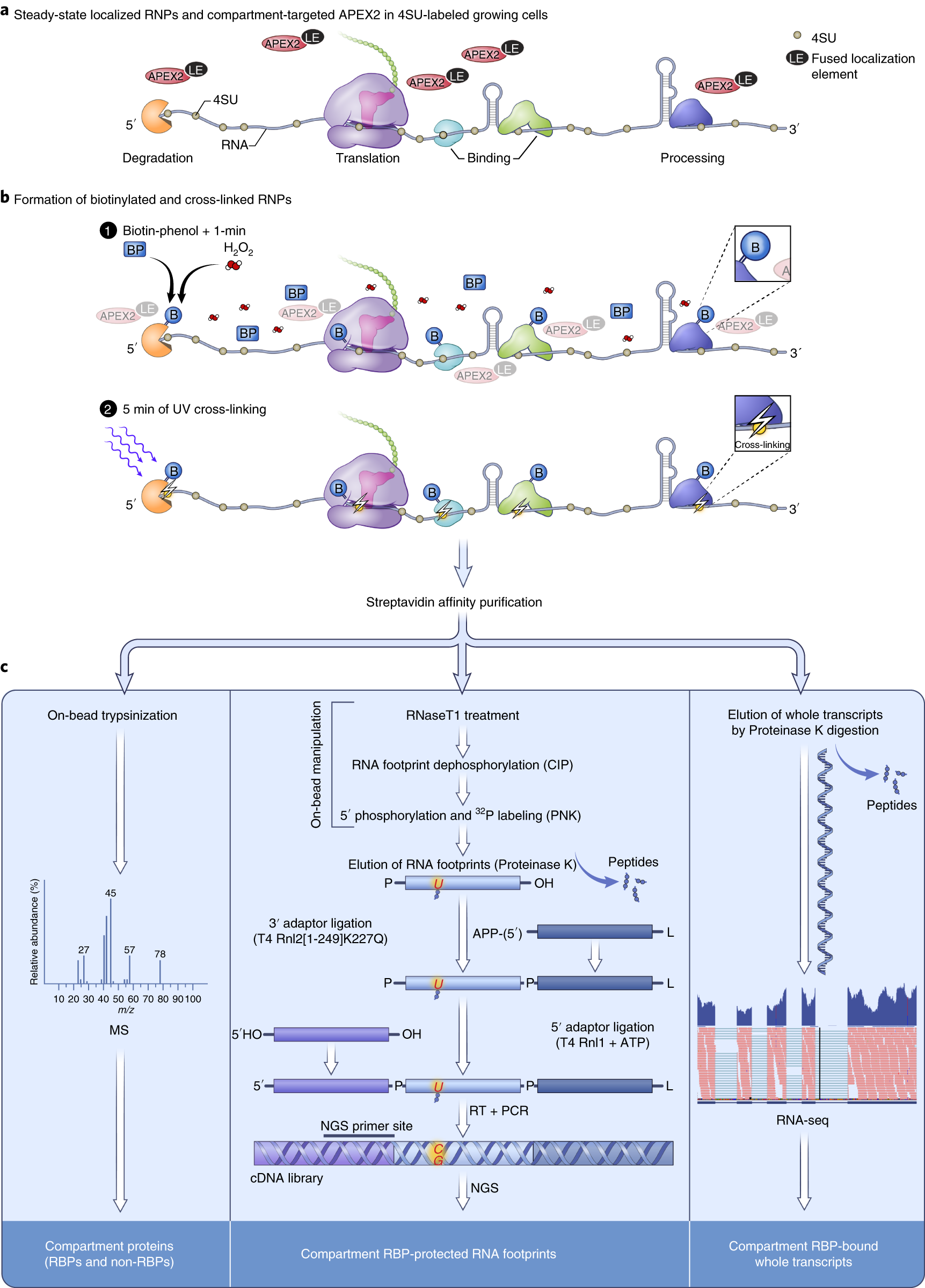
Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments | Nature Methods
![PDF] CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome | Semantic Scholar PDF] CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b403d8737a9f56397391232d083a351860a8215b/2-Figure1-1.png)
PDF] CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome | Semantic Scholar

MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein
Analysis of the nucleocytoplasmic shuttling RNA-binding protein HNRNPU using optimized HITS-CLIP method | PLOS ONE

Beyond CLIP: advances and opportunities to measure RBP-RNA and RNA-RNA interactions. - Abstract - Europe PMC

Global Approaches in Studying RNA-Binding Protein Interaction Networks: Trends in Biochemical Sciences
Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival
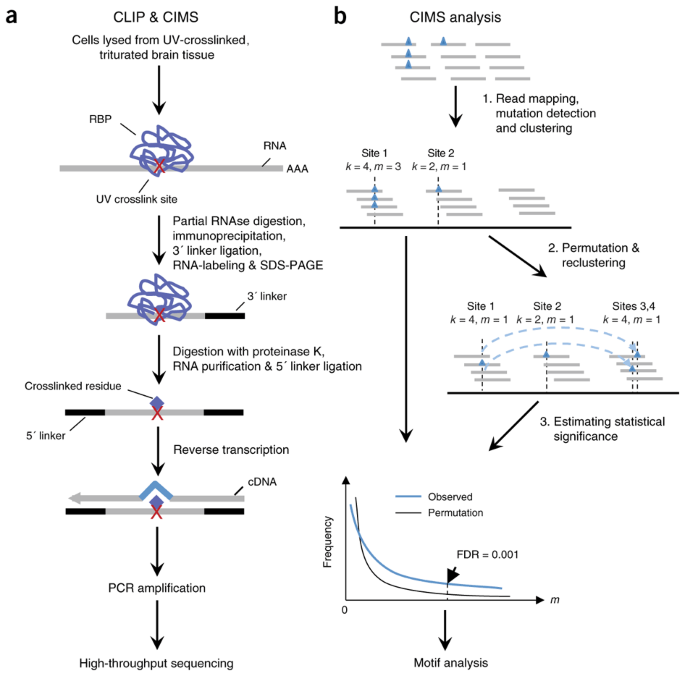
Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data | Nature Biotechnology

Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - ScienceDirect

omniCLIP: probabilistic identification of protein-RNA interactions from CLIP-seq data | RNA-Seq Blog
Protein – RNA interactions: Analysis of iCLIP-seq data FU Berlin Seminar RNA Bioinformatics, WS 14/15 Sabrina Krakau
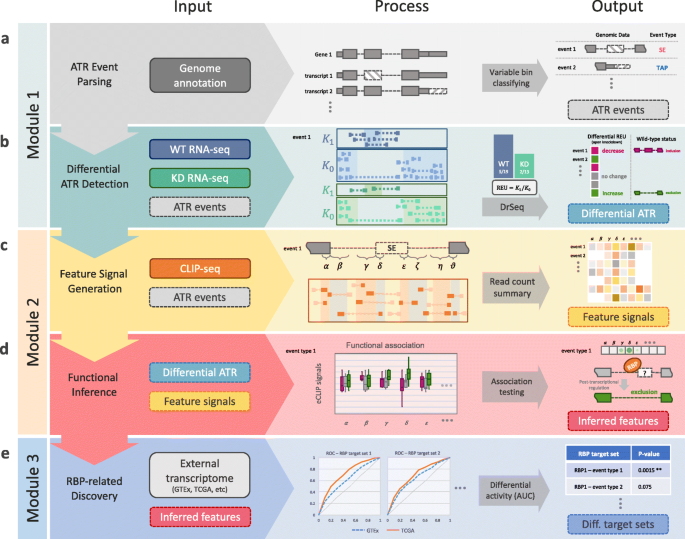
SURF: integrative analysis of a compendium of RNA-seq and CLIP-seq datasets highlights complex governing of alternative transcriptional regulation by RNA-binding proteins | Genome Biology | Full Text



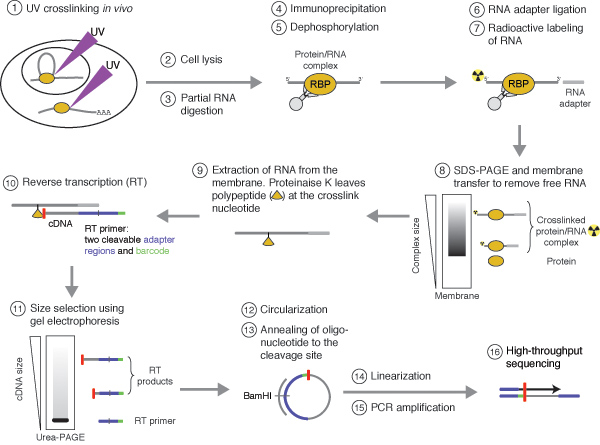
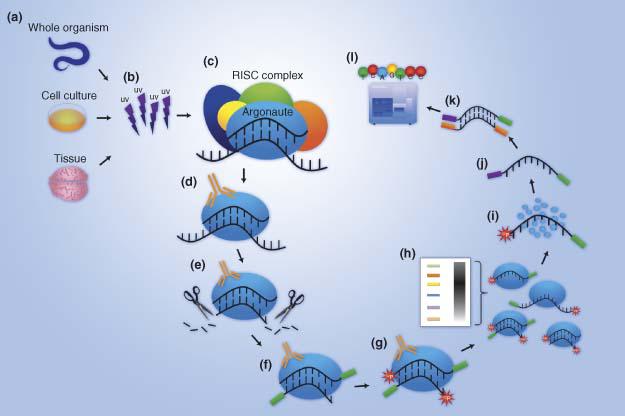
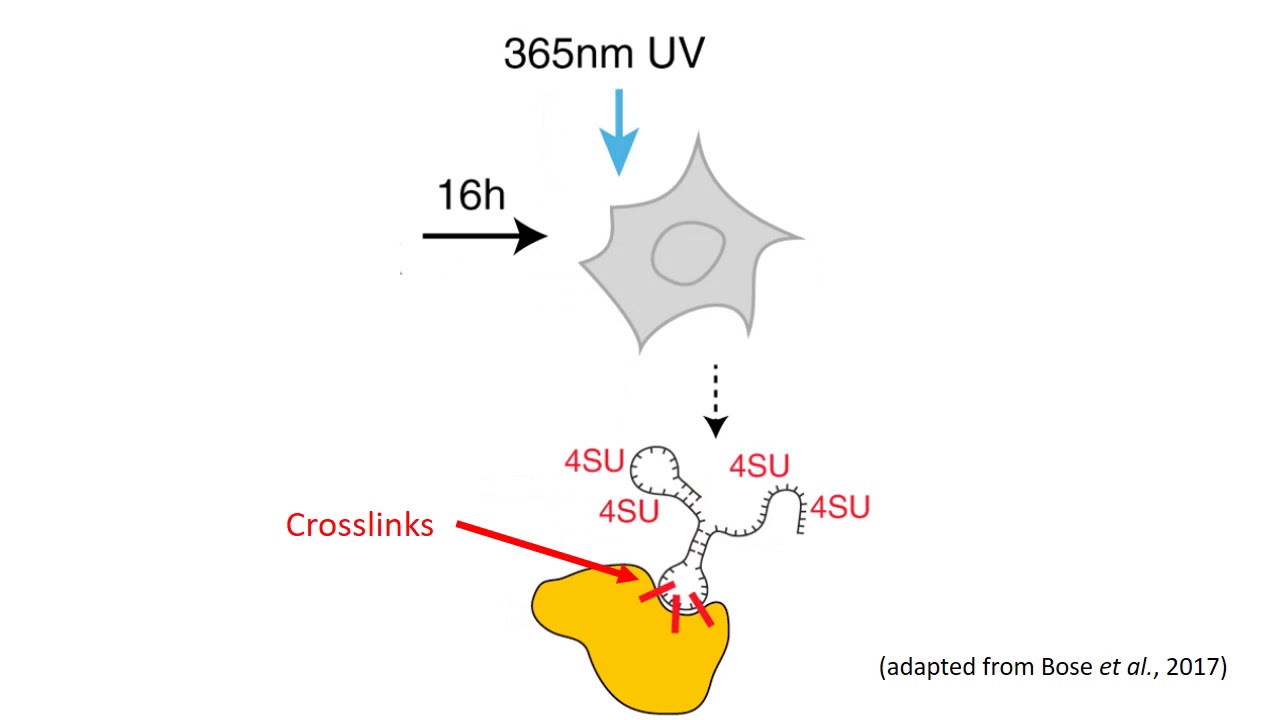
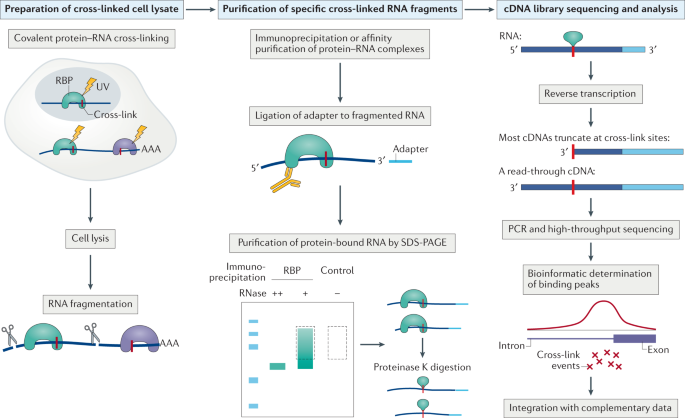


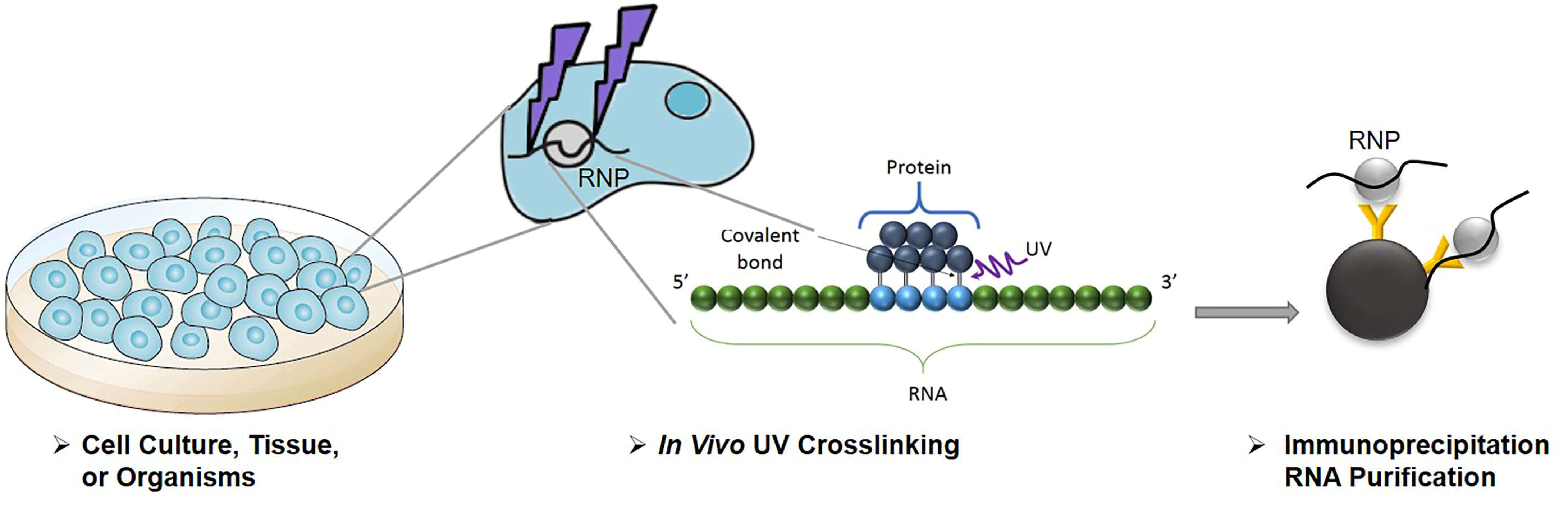
-2.jpg)